| 論文篇名 | 英文:Presence of an Agrobacterium-type tumor-inducing plasmid in Neorhizobium sp. NCHU2750 and the link to phytopathogenicity. 中文:在菌株Neorhizobium sp. NCHU2750中所含的腫瘤誘導質體及其致病性分析 |
| 期刊名稱 | Genome Biology and Evolution |
| 發表年份,卷數,起迄頁數 | 2018, 10, 3188-3195. |
| 作者 | Haryono, M., Tsai, Y.M., Lin, C.T., Huang, F.C., Ye, Y.C., Deng, W.L., Hwang, H.-H. (黃皓瑄),* Kuo, C.H.* |
| DOI | 10.1093/gbe/evy249 |
| 中文摘要 | 與中研院植微所郭志鴻研究員共同研究發現一株由台灣本土分離的新菌株Neorhizobium (NCHU2750),其菌體內含有類似農桿菌中的Ti質體,故可感染植物使其產生腫瘤。此研究並將其基因體解序分析後,其結果提供了根瘤菌科一個新的分類依據,並可用於未來更精確地分類根瘤菌科中的各種細菌,可作為比較基因體學中根瘤菌科分類依據的標準菌株。 |
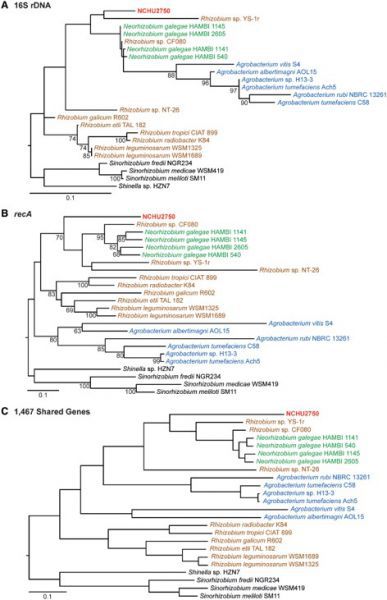
| 英文摘要 | The genus Agrobacterium contains a group of plant-pathogenic bacteria that have been developed into an important tool for genetic transformation of eukaryotes. To further improve this biotechnology application, a better understanding of the natural genetic variation is critical. During the process of isolation and characterization of wild-type strains, we found a novel strain (i.e., NCHU2750) that resembles Agrobacterium phenotypically but exhibits high sequence divergence in several marker genes. For more comprehensive characterization of this strain, we determined its complete genome sequence for comparative analysis and performed pathogenicity assays on plants. The results demonstrated that this strain is closely related to Neorhizobium in chromosomal organization, gene content, and molecular phylogeny. However, unlike the characterized species within Neorhizobium, which all form root nodules with legume hosts and are potentially nitrogen fixing mutualists, NCHU2750 is a gall-forming pathogen capable of infecting plant hosts across multiple families. Intriguingly, this pathogenicity phenotype could be attributed to the presence of an Agrobacterium-type tumor-inducing plasmid in the genome of NCHU2750. These findings suggest that these different lineages within the family Rhizobiaceae are capable of transitioning between ecological niches by having novel combinations of replicons. In summary, this work expanded the genomic resources available within Rhizobiaceae and provided a strong foundation for future studies of this novel lineage. With an infectivity profile that is different from several representative Agrobacterium strains, this strain may be useful for comparative analysis to better investigate the genetic determinants of host range among these bacteria. |
Presence of an Agrobacterium-type tumor-inducing plasmid in Neorhizobium sp. NCHU2750 the link to phytopathogenicity 2018-11-06
循環農業:農業廢棄物再資源化【生科系黃皓瑄副教授】